[8]:
import logging
import h5py
import dill
import matplotlib.pyplot as plt
import numpy as np
import scipy.stats as stats
import SpectralToolbox.Spectral1D as S1D
import TransportMaps as TM
import TransportMaps.Maps.Functionals as FUNC
import TransportMaps.Maps as MAPS
import TransportMaps.Distributions as DIST
from TransportMaps import KL
TM.setLogLevel(logging.INFO)
Sparse transports for Markov random fields¶
Markov properties of the target distribution \(\nu_\pi\) can be exploited in order to derive efficient low-dimensional approximations [TR4]. We alreadi discussed (here) the decomposability of direct transports \(T\). Here we present explore the sparsity of the inverse transport \(S\).
We use again the stochastic volatility model as an example. See [OR7] and [OR8] for a presentation of the problem. We use the solution to the sequential inference problem to create a synthetic example, i.e. to generate a sample for which we will assume not to know its origin.
Let us load such a sample (we use a \(10^4\) long Markov chain from \(\nu_\pi\)) …
[3]:
f = h5py.File("auxiliary/inv-stocvol/Sequential-IntSq-Postprocess.dill.hdf5")
x = f['metropolis-independent-proposal-samples']['x'][:]
w = np.ones(x.shape[0]) / float(x.shape[0])
f.close()
Let us then define the distribution \(\nu_\pi\) …
[4]:
class StocVol(DIST.Distribution):
def __init__(self, x):
super(StocVol,self).__init__(x.shape[1])
self.x = x
def quadrature(self, qtype, qparams, *args, **kwargs):
if qtype == 1: # We use 1 to indicate a Markov chain
nmax = self.x.shape[0]
if qparams > nmax:
raise ValueError("Maximum sample size (%d) exceeded" % nmax)
x = self.x[:qparams,:]
w = np.ones(qparams)/float(qparams)
else: raise ValueError("Quadrature not defined")
return (x,w)
pi = StocVol(x)
Let us now define a simple linear transformation that rescales the position of the sample to the hyper-cube \([-4,4]\) (as in the introductory example) …
[5]:
a = np.zeros(pi.dim)
b = np.zeros(pi.dim)
for d in range(pi.dim):
xmax = np.max(x[:,d])
xmin = np.min(x[:,d])
a[d] = 4*(xmin+xmax)/(xmin-xmax)
b[d] = 8./(xmax-xmin)
L = MAPS.FrozenLinearDiagonalTransportMap(a, b)
Sparse inverse transports¶
We know by construction that the stochastic volatility model will have the following Markov structure:
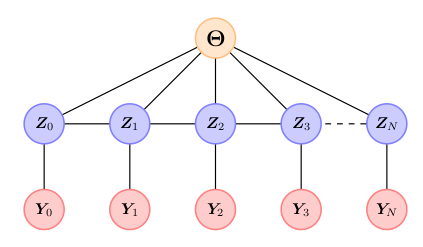
where \(\Theta=(\mu,\phi)\) and \({\bf Z}_k \in \mathbb{R}\). Using the theory on sparse inverse transports [TR4] we then expect \(S\) to have the following sparsity structure:
Let us then assemble the map \(S\) of total order 3.
[7]:
order = 2
active_vars = [[0], [0,1], [0,1,2], [0,1,2,3],
[0,1,3,4], [0,1,4,5]]
S = MAPS.assemble_IsotropicIntegratedSquaredTriangularTransportMap(
6, order, 'total', active_vars=active_vars)
print("Number of coefficients: %d" % S.n_coeffs)
Number of coefficients: 64
Variational solution¶
We can now set up the problem
where \(\mathcal{T}_\triangle\) is the set of integrated squared triangular transport maps (presented here) of (total) order 3 and with the aforementioned sparsity pattern.
[9]:
rho = DIST.StandardNormalDistribution(6)
push_L_pi = DIST.PushForwardTransportMapDistribution(L, pi)
push_SL_pi = DIST.PushForwardTransportMapDistribution(
S, push_L_pi)
pull_rho = DIST.PullBackParametricTransportMapDistribution(
S, rho)
qtype = 1 # Monte-Carlo quadratures from pi
qparams = 5000 # Number of MC points
reg = {'type': 'L2',
'alpha': 1e-2} # L2 regularization
tol = 1e-3 # Optimization tolerance
ders = 2 # Use gradient and Hessian
log = KL.minimize_kl_divergence(
push_L_pi, pull_rho, qtype=qtype, qparams=qparams, regularization=reg,
tol=tol, ders=ders)
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Optimization failed.
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Message: Warning: Desired error not necessarily achieved due to precision loss.
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Function value: 1.219170
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Norm of the Jacobian: 0.009013
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Number of iterations: 3
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. function evaluations: 27
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Jacobian evaluations: 15
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Hessian evaluations: 4
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Optimization failed.
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Message: Warning: Desired error not necessarily achieved due to precision loss.
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Function value: 1.325452
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Norm of the Jacobian: 0.000062
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Number of iterations: 4
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. function evaluations: 21
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Jacobian evaluations: 9
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Hessian evaluations: 5
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Optimization failed.
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Message: Warning: Desired error not necessarily achieved due to precision loss.
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Function value: 1.262508
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Norm of the Jacobian: 0.001154
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Number of iterations: 5
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. function evaluations: 25
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Jacobian evaluations: 13
2023-03-26 01:06:05 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Hessian evaluations: 6
2023-03-26 01:06:06 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Optimization terminated successfully
2023-03-26 01:06:06 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Function value: 1.175963
2023-03-26 01:06:06 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Norm of the Jacobian: 0.000079
2023-03-26 01:06:06 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Number of iterations: 8
2023-03-26 01:06:06 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. function evaluations: 9
2023-03-26 01:06:06 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Jacobian evaluations: 9
2023-03-26 01:06:06 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Hessian evaluations: 8
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Optimization terminated successfully
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Function value: 1.155457
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Norm of the Jacobian: 0.000009
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Number of iterations: 9
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. function evaluations: 10
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Jacobian evaluations: 10
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Hessian evaluations: 9
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Optimization terminated successfully
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Function value: 1.199731
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Norm of the Jacobian: 0.000053
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: Number of iterations: 8
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. function evaluations: 9
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Jacobian evaluations: 9
2023-03-26 01:06:07 INFO: TM.IntegratedSquaredParametricMonotoneFunctional: N. Hessian evaluations: 8
Note that the solutioon of this variational problem does not require any evaluation of the density \(\pi\), but only evaluations of \(\rho\), which is trivial.
Accuracy of the approximation¶
In general the accuracy of this approximation cannot be evaluated as with the diagnostics presented for the direct approach, due to the un-availability of the density \(\pi\).
For the sake of the presentation, though, we will compare the approximation \(L^\sharp S^\sharp \rho\) against the actual density \(\pi\), to check its accouracy.
[10]:
with open("auxiliary/inv-stocvol/Distribution.dill", 'rb') as in_stream:
pi_star = dill.load(in_stream)
Variance diagnostic¶
We compute the quantity \(\mathbb{V}{\rm ar}_\pi \left( \log \frac{L^\sharp S^\sharp \rho}{\pi} \right)\) …
[11]:
import TransportMaps.Diagnostics as DIAG
SL = MAPS.CompositeTransportMap(S, L)
pull_SL_rho = DIST.PullBackTransportMapDistribution(SL, rho)
var = DIAG.variance_approx_kl(pull_SL_rho, pi_star, x=x, w=w)
print("Variance diagnostic: %e" % var)
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Maps/MapBase.py:57, in Map.dim_in(self)
56 try:
---> 57 self._dim_in
58 except AttributeError:
59 # backward compatibility
AttributeError: 'IndependentLogLikelihood' object has no attribute '_dim_in'
During handling of the above exception, another exception occurred:
KeyError Traceback (most recent call last)
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Maps/MapBase.py:60, in Map.dim_in(self)
58 except AttributeError:
59 # backward compatibility
---> 60 self._dim_in = vars(self)['dim_in']
61 del vars(self)['dim_in']
KeyError: 'dim_in'
During handling of the above exception, another exception occurred:
AttributeError Traceback (most recent call last)
Cell In[11], line 4
2 SL = MAPS.CompositeTransportMap(S, L)
3 pull_SL_rho = DIST.PullBackTransportMapDistribution(SL, rho)
----> 4 var = DIAG.variance_approx_kl(pull_SL_rho, pi_star, x=x, w=w)
5 print("Variance diagnostic: %e" % var)
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Diagnostics/Routines.py:157, in variance_approx_kl(d1, d2, params1, params2, vals_d1, vals_d2, qtype, qparams, x, w, mpi_pool_tuple, import_set)
153 elif x is None or w is None:
154 raise ValueError("Provide quadrature points and weights or quadrature " + \
155 "type and parameters")
--> 157 vals_d1, vals_d2 = compute_vals_variance_approx_kl(
158 d1, d2, params1, params2, x=x,
159 mpi_pool_tuple=mpi_pool_tuple, import_set=import_set)
161 vals = vals_d1 - vals_d2
162 expect = np.dot( vals, w )
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Diagnostics/Routines.py:52, in compute_vals_variance_approx_kl(d1, d2, params1, params2, x, mpi_pool_tuple, import_set)
49 vals_d1 = mpi_map("log_pdf", obj=d1, scatter_tuple=scatter_tuple,
50 mpi_pool=mpi_pool_tuple[0])
51 # d2
---> 52 vals_d2 = mpi_map("log_pdf", obj=d2, scatter_tuple=scatter_tuple,
53 mpi_pool=mpi_pool_tuple[1])
54 return (vals_d1, vals_d2)
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/MPI.py:204, in mpi_map(f, scatter_tuple, bcast_tuple, dmem_key_in_list, dmem_arg_in_list, dmem_val_in_list, obj, obj_val, reduce_obj, reduce_tuple, mpi_pool, splitted, concatenate)
202 f = getattr(obj, f)
203 # Evaluate
--> 204 fval = f(**args)
205 # Reduce if necessary
206 if reduce_obj is not None:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:272, in cached.__call__.<locals>.wrapped(slf, *args, **kwargs)
270 raise TypeError("Type of sub_len not recognized")
271 # Evaluate function
--> 272 out = f(slf, *args, **kwargs)
273 # Store in cache
274 if caching:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:133, in counted.<locals>.wrapped(slf, *args, **kwargs)
131 slf.nevals[f.__name__] = x.shape[0]
132 start = process_time()
--> 133 out = f(slf, *args, **kwargs)
134 stop = process_time()
135 try:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Distributions/Inference/InferenceBase.py:112, in BayesPosteriorDistribution.log_pdf(self, x, idxs_slice, cache, **kwargs)
98 r""" Evaluate :math:`\log \pi({\bf x}\vert{\bf y})`
99
100 Args:
(...)
109 at the ``x`` points.
110 """
111 logL_cache, prior_cache = get_sub_cache(cache, ("logL",None), ("prior",None))
--> 112 return self.logL.evaluate(x, idxs_slice=idxs_slice, cache=logL_cache)[:,0] \
113 + self.prior.log_pdf(x, idxs_slice=idxs_slice, cache=prior_cache)
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:272, in cached.__call__.<locals>.wrapped(slf, *args, **kwargs)
270 raise TypeError("Type of sub_len not recognized")
271 # Evaluate function
--> 272 out = f(slf, *args, **kwargs)
273 # Store in cache
274 if caching:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:133, in counted.<locals>.wrapped(slf, *args, **kwargs)
131 slf.nevals[f.__name__] = x.shape[0]
132 start = process_time()
--> 133 out = f(slf, *args, **kwargs)
134 stop = process_time()
135 try:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Likelihoods/LikelihoodBase.py:755, in IndependentLogLikelihood.evaluate(self, x, idxs_slice, cache, **kwargs)
743 @cached([("ll_list","n_factors")])
744 @counted
745 def evaluate(self, x, idxs_slice=slice(None,None,None), cache=None, **kwargs):
746 r""" Evaluate :math:`\log\pi({\bf y} \vert {\bf x})`.
747
748 Args:
(...)
753 (:class:`ndarray<numpy.ndarray>` [:math:`m,1`]) -- function evaluations
754 """
--> 755 if x.shape[1] != self.dim_in:
756 raise ValueError("The dimension of the input does not match the dimension " + \
757 "of the log-likelihood")
758 ll_list_cache = get_sub_cache(cache, ("ll_list",self.n_factors))
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Maps/MapBase.py:63, in Map.dim_in(self)
61 del vars(self)['dim_in']
62 finally:
---> 63 return self._dim_in
AttributeError: 'IndependentLogLikelihood' object has no attribute '_dim_in'
PushForward conditionals¶
We can plot slices of \(S_\sharp L_\sharp \nu_\pi \approx \nu_\rho \sim \mathcal{N}(0,{\bf I})\) …
[12]:
push_SL_pi_star = DIST.PushForwardTransportMapDistribution(SL, pi_star)
fig = DIAG.plotAlignedConditionals( push_SL_pi_star, range_vec=[-5,5],
numPointsXax=20, fig=plt.figure(figsize=(6.5,6.5)), show_flag=False,
vartitles=[r"$\mu$",r"$\phi$",r"${\bf Z}_0$",r"${\bf Z}_1$",r"${\bf Z}_2$",r"${\bf Z}_3$"])
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Maps/MapBase.py:57, in Map.dim_in(self)
56 try:
---> 57 self._dim_in
58 except AttributeError:
59 # backward compatibility
AttributeError: 'IndependentLogLikelihood' object has no attribute '_dim_in'
During handling of the above exception, another exception occurred:
KeyError Traceback (most recent call last)
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Maps/MapBase.py:60, in Map.dim_in(self)
58 except AttributeError:
59 # backward compatibility
---> 60 self._dim_in = vars(self)['dim_in']
61 del vars(self)['dim_in']
KeyError: 'dim_in'
During handling of the above exception, another exception occurred:
AttributeError Traceback (most recent call last)
Cell In[12], line 2
1 push_SL_pi_star = DIST.PushForwardTransportMapDistribution(SL, pi_star)
----> 2 fig = DIAG.plotAlignedConditionals( push_SL_pi_star, range_vec=[-5,5],
3 numPointsXax=20, fig=plt.figure(figsize=(6.5,6.5)), show_flag=False,
4 vartitles=[r"$\mu$",r"$\phi$",r"${\bf Z}_0$",r"${\bf Z}_1$",r"${\bf Z}_2$",r"${\bf Z}_3$"])
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Diagnostics/Plotting.py:185, in plotAlignedConditionals(distribution, data, dimensions_vec, range_vec, numPointsXax, numCont, figname, show_flag, do_diag, show_title, show_axis, title, vartitles, fig, ret_handles, mpi_pool)
182 raise RuntimeError("Plotting is not suported")
184 if data is None:
--> 185 data = computeAlignedConditionals(
186 distribution, dimensions_vec=dimensions_vec,
187 range_vec=range_vec, numPointsXax=numPointsXax,
188 mpi_pool=mpi_pool)
189 nplots = data.nplots
190 X_list = data.X_list
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Diagnostics/Plotting.py:136, in computeAlignedConditionals(distribution, dimensions_vec, range_vec, numPointsXax, do_diag, mpi_pool)
134 tempMat_mat = np.concatenate(tempMat_list, axis=0)
135 scatter_tuple = (['x'], [tempMat_mat])
--> 136 pdfEval_mat = mpi_map("pdf", obj=distribution, scatter_tuple=scatter_tuple,
137 mpi_pool=mpi_pool)
138 pdfEval_list = [ pdfEval_mat[npoints_pos[i]:npoints_pos[i+1]]
139 for i in range(len(tempMat_list)) ]
141 data = AlignedConditionalsObject(nplots, X_list, Y_list, pdfEval_list)
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/MPI.py:204, in mpi_map(f, scatter_tuple, bcast_tuple, dmem_key_in_list, dmem_arg_in_list, dmem_val_in_list, obj, obj_val, reduce_obj, reduce_tuple, mpi_pool, splitted, concatenate)
202 f = getattr(obj, f)
203 # Evaluate
--> 204 fval = f(**args)
205 # Reduce if necessary
206 if reduce_obj is not None:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:133, in counted.<locals>.wrapped(slf, *args, **kwargs)
131 slf.nevals[f.__name__] = x.shape[0]
132 start = process_time()
--> 133 out = f(slf, *args, **kwargs)
134 stop = process_time()
135 try:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Distributions/TransportMapDistributions.py:69, in PushForwardTransportMapDistribution.pdf(self, x, params, idxs_slice, cache)
53 @counted
54 def pdf(self, x, params=None, idxs_slice=slice(None), cache=None):
55 r""" Evaluate :math:`T_\sharp \pi({\bf x})`
56
57 Args:
(...)
67 at the ``x`` points.
68 """
---> 69 return np.exp( self.log_pdf(x, params, idxs_slice=idxs_slice, cache=cache) )
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:272, in cached.__call__.<locals>.wrapped(slf, *args, **kwargs)
270 raise TypeError("Type of sub_len not recognized")
271 # Evaluate function
--> 272 out = f(slf, *args, **kwargs)
273 # Store in cache
274 if caching:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:133, in counted.<locals>.wrapped(slf, *args, **kwargs)
131 slf.nevals[f.__name__] = x.shape[0]
132 start = process_time()
--> 133 out = f(slf, *args, **kwargs)
134 stop = process_time()
135 try:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Distributions/TransportMapDistributions.py:105, in PushForwardTransportMapDistribution.log_pdf(self, x, params, idxs_slice, cache)
103 params_t['xinv'] = xinv
104 ldgx = self.transport_map.log_det_grad_x_inverse(x, precomp=params_t, idxs_slice=idxs_slice)
--> 105 lpdf = self.base_distribution.log_pdf(xinv, params=params_pi)
106 return TransportMapDistribution._evaluate_log_transport(lpdf, ldgx)
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:272, in cached.__call__.<locals>.wrapped(slf, *args, **kwargs)
270 raise TypeError("Type of sub_len not recognized")
271 # Evaluate function
--> 272 out = f(slf, *args, **kwargs)
273 # Store in cache
274 if caching:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:133, in counted.<locals>.wrapped(slf, *args, **kwargs)
131 slf.nevals[f.__name__] = x.shape[0]
132 start = process_time()
--> 133 out = f(slf, *args, **kwargs)
134 stop = process_time()
135 try:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Distributions/Inference/InferenceBase.py:112, in BayesPosteriorDistribution.log_pdf(self, x, idxs_slice, cache, **kwargs)
98 r""" Evaluate :math:`\log \pi({\bf x}\vert{\bf y})`
99
100 Args:
(...)
109 at the ``x`` points.
110 """
111 logL_cache, prior_cache = get_sub_cache(cache, ("logL",None), ("prior",None))
--> 112 return self.logL.evaluate(x, idxs_slice=idxs_slice, cache=logL_cache)[:,0] \
113 + self.prior.log_pdf(x, idxs_slice=idxs_slice, cache=prior_cache)
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:272, in cached.__call__.<locals>.wrapped(slf, *args, **kwargs)
270 raise TypeError("Type of sub_len not recognized")
271 # Evaluate function
--> 272 out = f(slf, *args, **kwargs)
273 # Store in cache
274 if caching:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Misc.py:133, in counted.<locals>.wrapped(slf, *args, **kwargs)
131 slf.nevals[f.__name__] = x.shape[0]
132 start = process_time()
--> 133 out = f(slf, *args, **kwargs)
134 stop = process_time()
135 try:
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Likelihoods/LikelihoodBase.py:755, in IndependentLogLikelihood.evaluate(self, x, idxs_slice, cache, **kwargs)
743 @cached([("ll_list","n_factors")])
744 @counted
745 def evaluate(self, x, idxs_slice=slice(None,None,None), cache=None, **kwargs):
746 r""" Evaluate :math:`\log\pi({\bf y} \vert {\bf x})`.
747
748 Args:
(...)
753 (:class:`ndarray<numpy.ndarray>` [:math:`m,1`]) -- function evaluations
754 """
--> 755 if x.shape[1] != self.dim_in:
756 raise ValueError("The dimension of the input does not match the dimension " + \
757 "of the log-likelihood")
758 ll_list_cache = get_sub_cache(cache, ("ll_list",self.n_factors))
File ~/.virtualenvs/transportmaps-private/lib/python3.10/site-packages/TransportMaps/Maps/MapBase.py:63, in Map.dim_in(self)
61 del vars(self)['dim_in']
62 finally:
---> 63 return self._dim_in
AttributeError: 'IndependentLogLikelihood' object has no attribute '_dim_in'
<Figure size 650x650 with 0 Axes>